
Hi, I'm Rubén Cañadas
My name is Rubén, and to be honest, I don't really know what I am.
I'm not a physicist.
I'm not a mathematician.
I'm not an AI engineer.
I'm not a software engineer.
Or maybe I'm a bit of all of them.
I like everything, I learn from everything, and I refuse to put myself in a box.
On a good day, we can talk about pseudo-Riemannian manifolds or tensor algebra.
On another, about how quantizing the Poisson bracket gives rise to commutators in quantum mechanics.
And if that feels too theoretical, we can jump straight into improving a multi-head attention mechanism or fine-tuning a Transformer with QLoRA.
Curiosity is the common thread.
That's why I've decided not to define myself on my own website.
I'm not a title. I'm not a label.
I'm just someone who's endlessly curious.
I could tell you about my career path, my degrees, my experience — all the usual stuff.
It would be perfectly fine.
But everyone does that.
That's what LinkedIn is for.
But anyway, here's what I've been focused on lately:
I'm bringing physics, mathematics, and artificial intelligence together in the world of computational biology.
The goal is not just to simulate molecules or predict structures.
It is to understand, in depth, how biology encodes information.
How physical laws, mathematical structure, and chemical constraints shape molecules and proteins.
How meaning emerges from form.
How function is written into matter.
Hidden inside molecular structures is an intrinsic language — a code shaped by evolution and physics.
By learning to read that code, we can build models that don't just predict outcomes, but understand.
Systems that capture structure, dynamics, and intent.
Systems that reason about biology, rather than approximate it.
That is the direction I'm exploring.
Things I'm Exploring Right Now
Topics and technologies I'm currently diving deep into
CUDA Kernels with Triton
Writing custom CUDA kernels using Triton to optimize inference, finetuning, and training pipelines. Achieving significant speedups in deep learning workloads.
Algebraic Topology for Molecular Docking
Exploring how topological data analysis and algebraic topology can help find optimal binding spaces for molecular docking simulations.
Preference Alignment (DPO, RLHF, RLAIF)
Studying and implementing preference alignment techniques like Direct Preference Optimization (DPO), RLHF, and RLAIF to better align language models with human preferences.
Ships & Builds
Turning ideas into profitable products that empower others to grow.
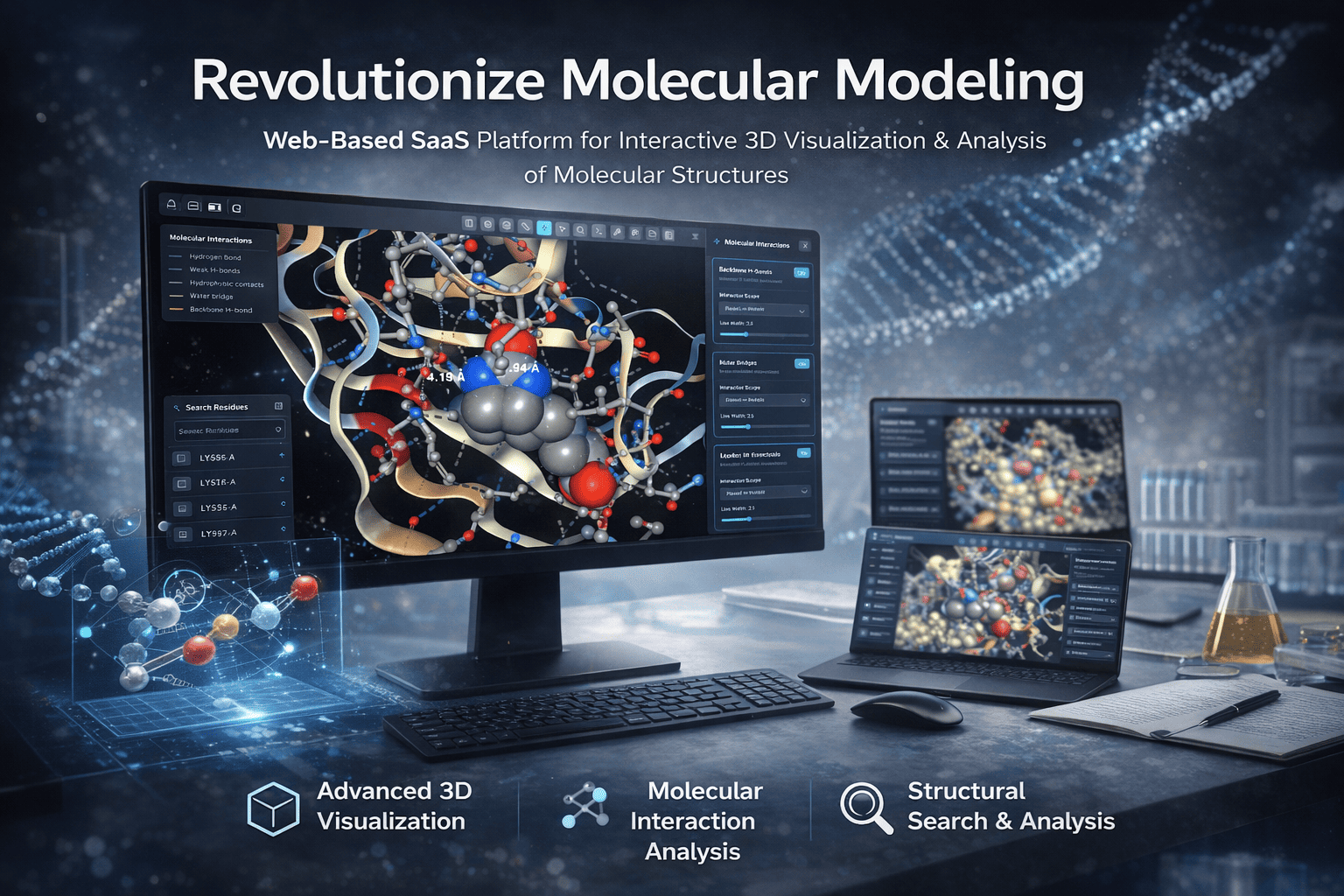
Nomosis - Protein Engineering Revolution
Revolutionary platform for protein engineering and drug design. Accelerates computational calculations x1000 by improving classic algorithms and adding AI accelerators. Calculations that took a month now take less than a day.
- 1000x faster calculations
- AI agents for hypothesis generation
- Experiment proposal system

Novachef - Recipe Revolution
Mobile app powered by artificial intelligence with 3000+ recipes. Features intelligent system for recipe questions, cooking tricks, shopping lists, menu creation, and AI chat agent expert in cooking.
- 3000+ recipes library
- AI-powered recipe assistant
- Shopping lists
Open source projects
Community contributions that aim to solve real problems and accelerate research.
MolFun
High-performance library to accelerate training and inference calculations for molecular modelling models like Protenix, AlphaFold and more.
10x faster
Low-level optimizations for molecular calculations
GPU Accelerated
Native support for CUDA and parallel operations
Easy integration
Compatible with PyTorch, JAX and popular frameworks
Molecular Modelling
Optimized for Protenix, AlphaFold, ESMFold
Articles & Tutorials
Thoughts & Deep Dives
Exploring the frontiers of AI, machine learning, and computational biology. From practical tutorials to deep technical insights.
 Machine Learning
Machine LearningUnderstanding Attention: The Idea Behind Modern AI
One paper changed everything: Attention Is All You Need. This post breaks down the foundations behind attention, starting from embeddings. A simple journey from words to meaning.
 Machine Learning
Machine LearningWhat Are Transformers in AI?
Learn what Transformers are, how they work, and why they power models like GPT and modern AI systems. A clear, beginner-friendly introduction.
Get notified about new posts and projects
No spam, unsubscribe anytime. Your email is safe with me.
Contact
Let's Connect
Have a project in mind, a question about my work, or just want to say hello? I'd love to hear from you.
Get in Touch
Have a question or want to work together?